DNA Oligonucleotides
Picoforce Measurements of Single DNA Interaction
Figure 1. The chemical attachment process of 50-mer DNA oligonucleotides involves self-assembled monolayers (SAMs) of DNA-functionalized dendrons. The dendron used in this study was 9-anthrylmethyl N-({[tris({2-[({tris-[(2-carboxyethoxy)methyl]methyl}amino)carbonyl]ethoxy}methyl)-methyl]amino}carbonyl)propyl carbamate. Sample courtesy of Prof. J.W Park (Dept. of Chemistry, POSTECH)
Using force spectroscopy, interactions between single molecules can be observed by monitoring the attractive or “jump-in” forces associated with molecular binding events. Molecular binding experiments presented by Jung et al., (2007) were repeated using the XE-100 under the guidance of Prof. Park. With the oligonucleotide sequences immobilized on the dendron-modified surface, the F-d curve shows association and dissociation of the DNA duplex. The force constant of the cantilever was calibrated in solution for every measurement by the thermal tune method, the variation of which is 12~15 pN/nm.
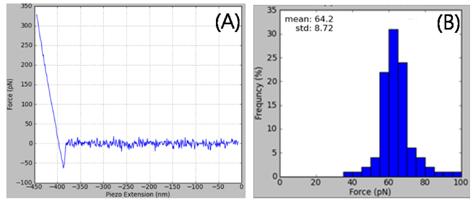
Figure 2. (A) A typical force-distance curve of the interaction between a DNA probe and the complementary 50-mer DNA. (B) The histogram of unbinding forces acquired from the force-distance curves. The mean value of 64 pN with a narrow distribution shows that AFM can be a useful tool to study the interactions between a single DNA molecule and other single DNA molecules, RNA molecules, and proteins.
Reference
Journal: J. Am. Chem. Soc. 2007, 129 (30), 9349–9355.
Title: Dendron Arrays for the Force-Based Detection of DNA Hybridization Events
Authors: Yu Jin Jung, Bong Jin Hong, Wenke Zhang, Saul J. B. Tendler, Philip M. Williams, Stephanie Allen, and Joon Won Park
