Single Strand DNA Molecules
NC-AFM Imaging of Bio-molecular Samples
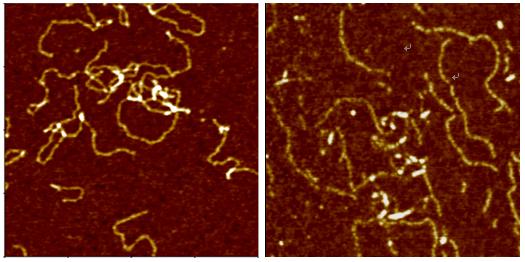
Deoxyribonucleic Acid (DNA) is a polymer chain that stores hereditary information of species. It is made from repeating units called nucleotides. In its natural state, the chain is 2.2 nm ~ 2.6 nm, and one nucleotide unit is 0.33 nm long. Although each individual repeating unit is very small, DNA polymers can be very large molecules containing millions of nucleotides. While X-ray crystallography and electron microscopy are the main tools used to resolve the structure of DNA in 3-dimensions, scanning probe techniques have gradually gained importance in the field of high resolution DNA imaging due to their ease of sample preparation (in-necessity to crystallize the DNA prior to imaging) as well as the possibility of functional studies (e.g., study of protein–DNA interaction processes) under physiological conditions. In preparation for AFM imaging, DNA samples are usually immobilized onto a stiff, flat substrate surface. The negatively charged backbone of the DNA can be utilized for immobilization onto charged substrates such as freshly cleaved mica via electrostatic interactions. After the DNA molecules are adsorbed onto the substrate, the surface can be washed, air-dried, and promptly imaged. Alternatively, the imaging process can be carried out in physiological solutions.
Publications Using the XE-AFM on DNA Molecules
Journal: Nucleic Acids Research, 33(20) November 2005. 6515.
Title: Synthesis of novel poly(dG)–poly(dG)–poly(dC) triplex structure by Klenow exofragment of DNA polymerase I
Authors: A. Kotlyar, N. Borovok1, T. Molotsky1, D. Klinov, B. Dwir and E. Kapon;
System: Park Systems XE-100
Abstract: The extension of the G-strand of long (700 bp) poly(dG)–poly(dC) by the Klenow exo-fragment of DNA polymerase I yields a complete triplex structure of the H-DNA type. Atomic force microscopy morphology imaging shows that the synthesized structures lack single-stranded fragments and have approximately the same length as the parent 700 bp poly(dG)–poly(dC). CD spectrum of the polymer has a large negative peak at 278 nm, which is characteristic of the poly(dG)–poly(dG)–poly(dC) triplex. The polymer is resistant to DNase and interacts much more weakly with ethidium bromide as compared with the double-stranded DNA.
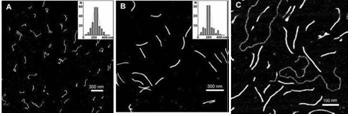
Fig. 2. AFM images of (A) synthesized poly(dG–dG)–poly(dC) molecules , (B) ~700 bp parent poly(dG)–poly(dC) and (C) poly(dG–dG)–poly(dC) molecules co-deposited with a linear plasmid pSK+ DNA on mica. Statistical length analyses of single, well-separated molecules from several images at several areas are shown in the inserts to (A) and (B).
