Gabriela Mendoza, Byong Kim and Keibock Lee, Park systems, INC., Santa Clara, CA, USA
Introduction
Probiotics are living microorganisms that provide numerous health benefits to the host when supplied in adequate amount as well as improve the intestinal microflora of the host. Probiotics are mainly administered to humans combined with food and are widely used in food products. From in vivo and in vitro studies of host intestinal epithelial or immune cell responses to probiotic strains, it has been reported that adhesion of probiotic strains to the intestinal surface and the subsequent colonization of the human gastrointestinal tract are important prerequisites for probiotic action [1]. This study looks into the topography and adhesion properties of those microorganisms to determine the shape and form that adheres well to the colon by means of using commercially available probiotics and the PinPoint nanomechanical mode of an atomic force microscopy (AFM) instrument from Park Systems.
Experimental
To isolate probiotic bacteria, 1 mL samples of a commercial product were put into microcentrifuge tubes and centrifuged (BioLab centrifuge) for 15 min at 10000 rpm to separate the biomass pellet. The supernatant was discarded and the biomass was recovered in a test tube. Serial dilutions (1 mL of biomass in 9 mL of sterile saline solution) from 10-1 to 10-5 were performed [2]. From the 10-5 dilutions, 0.1 mL drops were placed onto a silicon wafer chip (Ted Pella, Inc. 16007) and allowed to air dry. For bacteria characterization, a Park NX10 AFM from Park Systems was used; measurements were performed in air using an AC160TS cantilever. PinPoint™ Nanomechanical Mode from Park Systems can be used to study surface topography as well as determine quantitative nanomechanical properties (i.e., modulus, adhesion force, adhesion energy) [3]. In this study, topography, adhesion energy and adhesion force images were acquired using the PinPoint Nanomechanical Mode. Images obtained were 256 x 256 pixels and 2 μm x 2 μm sizes.
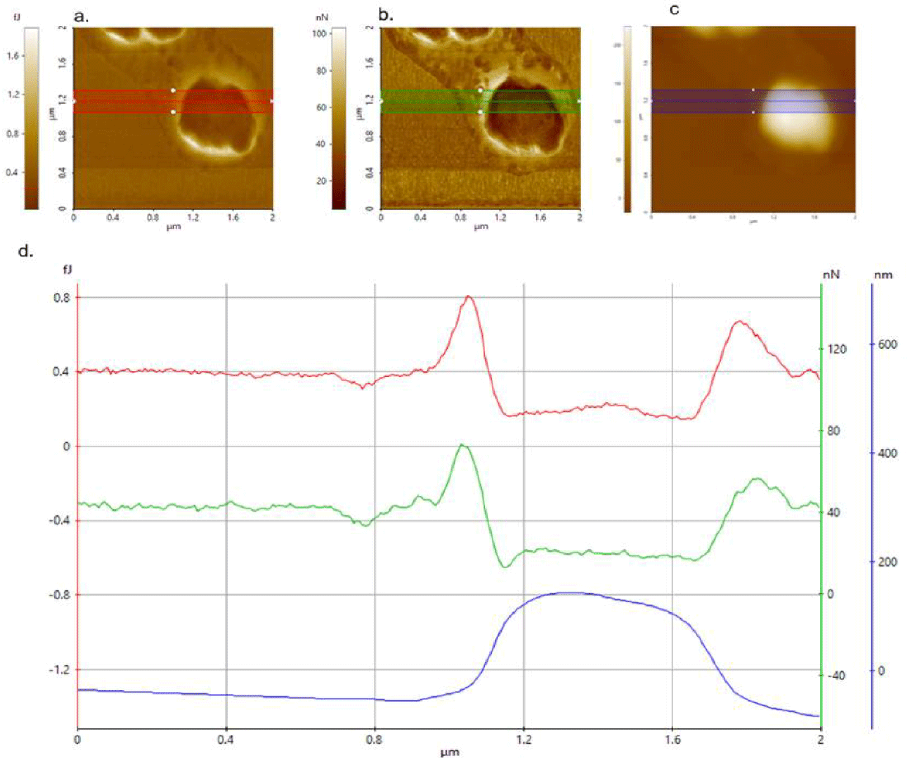
Figure 1. a) Adhesion energy, b) adhesion force and c) height images taken with AC160TS cantilever. d) Corresponding overlaid topography, adhesion energy and force line profiles.
Result
Figures 1a, b and c are the adhesion energy, adhesion force and topography images, respectively, of isolated Streptococcus chains from a 10-5 dilution. Figure 1d shows the corresponding overlaid topography, adhesion energy and force line profiles. The adhesion energy (Figure 1a) and adhesion force (Figure 1b) behaved in a similar manner; smaller values were observed for the bacteria compared with the substrate where they were attached.
The adhesion energy on the silicon wafer chip was 400 aJ, but just 200 aJ on the probiotic bacteria. Similar results were obtained for the adhesion force (Figure 1d); 20 nN for the bacteria but 45 nN on the silicon wafer chip and clearly indicates that less force was required on the bacteria. The isolated bacteria in air is observed to be relatively less adhesive than that of the underlying Si substrate. Bacterial adhesion is based on physical interactions between two or more surfaces. There were three different surfaces identified in this study: the cantilever tip (surface 1), the bacteria (surface 2) and the silicon wafer (surface 3). All three surfaces have different physicochemical properties: surface area, surface energy, surface roughness size and shape, which affect the adhesion force and energy.
Conclusion
The test results show that probiotic bacteria under study were less adhesive to the Si cantilever tip compared to the Si substrate. This phenomenon can be correlated with the adhesion of probiotic strains to the site of action, an important prerequisite for probiotic action. We found from nanomechanical measurements that the adhesion force and adhesion energy are affected by the physicochemical characteristics from each substrate. Therefore, adhesion force and adhesion energy values were lower when the measurement was on the bacteria compared to that done on the silicon wafer.
References
[1] Saarela M et al.,Journal of Biotechnology. 2000; (84) 197-215.
[2] Mendoza M AG et al., Revista Mexicana de Ingeniería Química. 2017; 16(1): 159-168.
[3] Park Systems’s PinPoint™ Nanomehcanical mode https://www.parksystems.com/index.php/en/parkafm- modes/nanomechanical-modes?i=0
