Biological recognition by force-distance spectroscopy, AFM
Research Application Technology Center
Introduction
Biological recognition is a fundamental process in biological systems that describes various phenomena that revolve around the characteristics of how molecules recognize each other and how specific and how strong their interactions are. Such molecules include typical components of biological systems such as proteins, peptides, lipids, and DNA. The governing forces between these species are usually non-covalent interactions like hydrophobic forces, hydrogen bonding, or ionic interactions.1 In most cases, the interactions between molecules are weak and non-specific. However, in the case of bio-recognition, two molecules can be complementary to each other like antibody and antigen pairs, forming complexes that are characterized by strong interactions, long lifetimes, and a specific binding affinity.1,2 In this regard, selectivity is the most important factor for bio-recognition, meaning that only one molecule responds strongly to a target molecule.
Being widely known as a versatile tool to image surface topographies, Atomic Force Microscopy (AFM) can also be used to investigate specific interactions in bio-recognition processes using a force-distance spectroscopy approach that allows studying mechanical properties (e.g., modulus, adhesion, and stiffness) of a sample. A force-distance (FD) plot shows the AFM tip and sample interaction as a function of tip-sample distance (Figure 1). In imaging mode, the tip scans over the sample surface to produce a three-dimensional (3D) image of the surface. In force-distance spectroscopy mode, the tip moves directly toward the sample until it establishes physical contact. Then, it retracts again to measure the interaction between the tip and the sample. To investigate the interaction between two partner molecules, they should be attached or immobilized onto the substrate and the AFM tip. Through FD curves obtained from force-distance spectroscopy, adhesion between two molecules can be measured and this AFM tip-sample interaction is utilized for studying the bio-recognition process3,4.
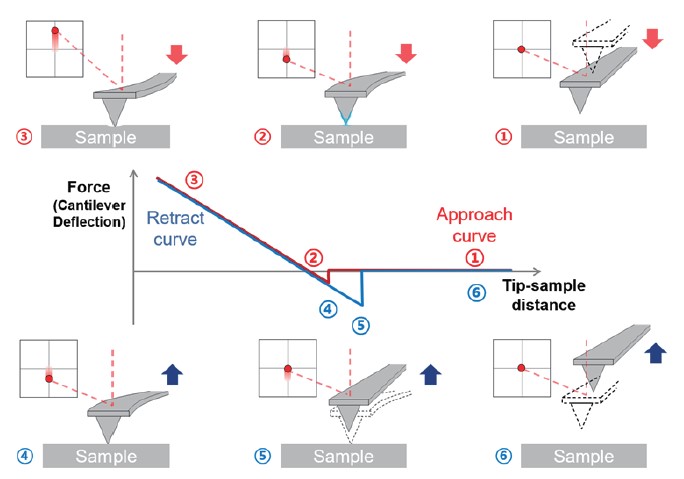
Figure 1. The procedure of recording FD curves. From the FD curve, mechanical properties can be obtained such as Young’s modulus, stiffness, adhesion force, and energy dissipation. Adhesion force is a mechanical and electrostatic force generated between different molecules. In AFM, it means the tip is trapped on the sample by force generated when the tip and the sample retract after contact.
In this application note, we introduce force-distance spectroscopy with functionalized AFM tips to measure specific interactions between nonstructural protein 5A (NS5A) amphipathic helical (AH) peptide and intracellular phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2). As shown in the literature, NS5A AH peptide specifically binds PI(4,5)P2 by a pair of positively charged lysine residues flanking a hydrophobic face5. Therefore, we investigate the possibility of using AFM to study the binding affinity of biomaterials (peptides) with extracellular matrix components.
AFM tip functionalization
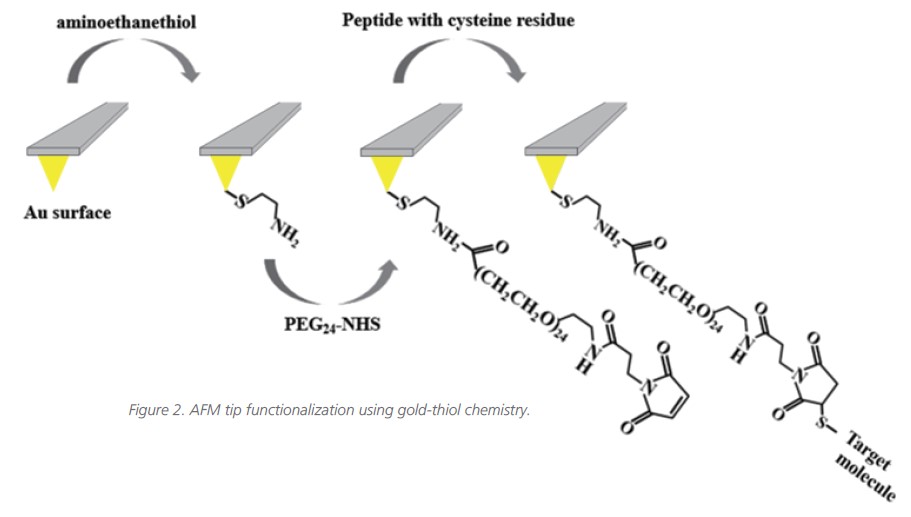
There are several methods to attach the functional organic molecule to the AFM tip, the most common approach is the formation of amino-self assembled monolayers (SAMs) at gold-coated AFM tip6. The main strong point of this method is a high binding affinity between thiol groups and gold. This high binding strength between gold and thiol groups can help to withstand unwanted removal of tip functionalization since the AFM tip-molecule interaction is stronger than the interaction between the tip-attached molecules and the molecules on the substrate. Moreover, gold-coated AFM tips can be easily reused by washing all immobilized molecules. For the measurement, we adopted gold-thiol chemistry for AFM tip functionalization. Because peptides like the one studied here are relatively small molecules with important secondary structure features, it is important to develop a chemical functionalization strategy that enables covalent attachment to the AFM tip while avoiding denaturation that would hinder binding.
Also, adopting the linker or spacer molecule enables the detection of specific binding events. When using non-functionalized tips or functionalization without a spacer, the results include many non-specific binding forces, and it is important to distinguish between non-specific interaction and the targeted specific interaction. The spacer between the AFM tip and molecule is typically flexible and it provides molecule mobility to easily access the binding of the surface molecule. It can characterize the rupture distance due to entropic stretching of the spacer molecule which can recognize the targeted specific interactions from non-specific interactions.
In this measurement, commercial rectangular-shaped soft AFM cantilevers (CSC38-Cantilever B, Mikromasch) with 0.03 N/m nominal spring constant were used to measure the binding interaction between peptide and phosphoinositide. At first, the AFM tips were coated with a 2.5 nm layer of titanium and then a 35 nm thick layer of gold (tip curvature radius is approximately 35 nm). For AFM tip functionalization, i.e., the peptide attachment, the tips were washed three times with ethanol solution and cleaned with a UV Ozone cleaner. The cleaning tips were incubated in ethanol solution including 2-aminoethanethiol (10mM) for 6 hours. After incubation, the tips were washed with a pure ethanol solution, dried with a very gentle stream of nitrogen gas, and immersed in an anhydrous toluene solution containing N-hydroxysuccinimide- PEG24-Maleimide ester (PEG24, 1 mmol/L) for 6 hours. For the final step, the immersed tips were rinsed with anhydrous toluene and ethanol and dried with a very gentle stream of nitrogen gas again. After that, the rinsed tips were incubated in PBS buffer solution with the target peptide overnight. Before conducting force-distance spectroscopy experiments, the peptide-attached tips were washed with PBS buffer solution (Figure 2). All measurements were performed by using the Park’s FX40 AFM, a new system that introduces automated tip handling, automated detection system alignment and tip access, and a multi-sample chuck.
Results
The sensing performance of the peptide-coated AFM tips is evaluated by conducting AFM force spectroscopic measurements on phosphoinositide- coated hydrophobic surfaces containing PI(4,5)P2 molecules. A representative FD curve on peptides-phosphoinositide is presented in figure 3. The black line shows a weak binding force (~200 pN) and the red line indicates a strong binding force (~400 pN) between two molecules. Both lines also show a rupture length of around 15 nm, which is consistent with the PEG spacer’s length of ~9.4 nm along with the position of the cysteine residue relative to the binding domain (~2 nm) and some unfolding of the peptide’s helical character during the unfolding (~3 nm)7,8. By contrast, when the AFM tip is not functionalized with peptides, there is no interaction (no adhesion force and rupture length) between the peptide and PI(4,5)P2 molecules (blue line), indicating that the detected interactions (black and red lines) are specific interactions.
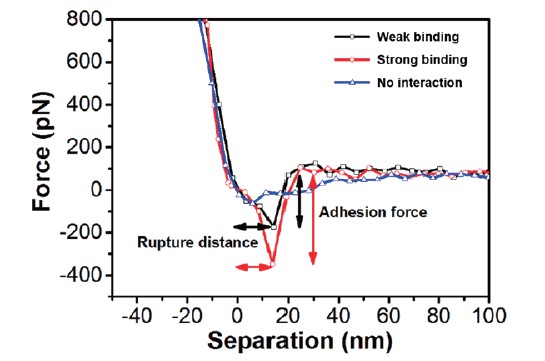
Figure 3. The FD curves (retract curve) of peptide attached AFM tip with partner molecules. Black and red lines show weak and strong binding respectively, and the blue line indicates no binding (interaction).
Next, force mapping measurement is performed. For force mapping data-set acquisition, the mapping resolution was 16 × 16 points with 10 × 10 μm2 of the scanning area, resulting in 256 FD curves per one dataset. In order to accurately analyze the statistical data, a large number of interacting molecules must be investigated in each case since biomolecular complexes contain various variables. As shown in figure 4, the statistical distribution of the adhesion force and the rupture length is calculated from the FD curves. In the case of no peptide attachment on the AFM tip, the averaged adhesion force has tiny values (~40 pN) indicative of weak, non-specific adhesion events. By contrast, in the case of peptide-attached AFM tips, the most probable adhesion force of peptide-PI(4,5)P2 interaction indicates around 400 pN. The statistical data clearly show the difference between the AFM tip with and without peptide, which shows the possibility of studying the specific or non-specific interaction of biomaterials using AFM force-distance spectroscopy.
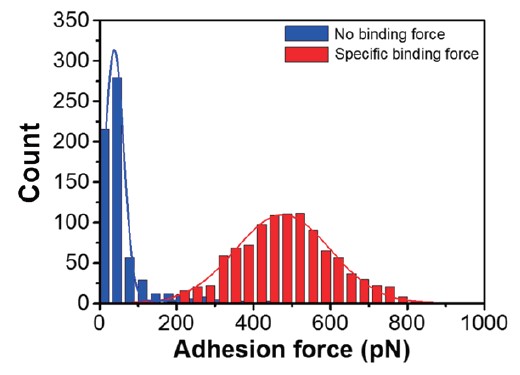
Figure 4. Binding force distributions of events with no interaction and specific interaction. Blue graph: No interaction, red graph: Specific binding events between two molecules. The histograms are fitted to Gaussian distribution to recognize the most probable binding force values.
Conclusion
We have introduced an experimental approach based on AFM force-distance spectroscopy that is able to measure the adhesion force associated with specific binding interactions between peptide and phosphoinositide. The experimental approach is based on covalently tethering the peptide to a functionalized, gold-coated AFM probe tip. The binding affinity between the two molecules was confirmed by attaching peptides to the AFM tip and using force-distance spectroscopy on phosphoinositide that specifically interacts with the peptide. In addition, for clear comparison, non-specific interaction was confirmed by measuring phosphoinositide with an AFM tip to which no peptide was attached.
An AFM is a useful technique to image the surface of a material, but it can also confirm specific binding between biomaterials as shown in this application note. The outcome of this work suggests the potential of AFM in studying the interaction between proteins, peptides, other biomaterials, and inter or extra-cellular components, and identifying intracellular or intercellular signaling pathways.
References
1. Wilchek, M., Bayer, E. A. & Livnah, O. Essentials of biorecognition: the (strept) avidin–biotin system as a model for protein–protein and protein–ligand interaction. Immunology letters 103, 27-32 (2006).
2. Noy, A. Handbook of molecular force-distance spectroscopy. (Springer Science & Business Media, 2007).
3. Dupres, V., Verbelen, C. & Dufrêne, Y. F. Probing molecular recognition sites on biosurfaces using AFM. Biomaterials 28, 2393-2402 (2007).
4. Ha, T. & Selvin, P. R. Single-molecule techniques: A laboratory manual. (Cold Spring Harbor Laboratory Press, 2008).
5. Cho, N.-J. et al. Phosphatidylinositol 4, 5-bisphosphate is an HCV NS5A ligand and mediates replication of the viral genome. Gastroenterology 148, 616-625 (2015).
6. Ebner, A. et al. in STM and AFM Studies on (Bio) Molecular Systems: Unravelling the Nanoworld 29-76 (Springer, 2008).
7. Lantz, M. A. et al. Stretching the α-helix: a direct measure of the hydrogen-bond energy of a single-peptide molecule. Chemical Physics Letters 315, 61-68 (1999).
8. Arai, Y., Okabe, K.-I., Sekiguchi, H., Hayashi, T. & Hara, M. Nanoscale chemical composition analysis using peptides targeting inorganic materials. Langmuir 27, 2478-2483 (2011).
